Cool Tools: Visualizing the Invisible
From fluorescence imaging that lights up proteins to electron microscopy that pinpoints cellular structures, visualization techniques have literally and figuratively illuminated the inner workings of cells. Still, the function of many proteins and molecules within cells as well as the details of certain critical cellular processes remain hard to see.
Thanks in part to research funded by the National Institutes of Health, the ability to peer further into the body at the cellular and subcellular levels is improving our understanding of cell structure, function and regulation—all of which are involved in both normal and disease states.
Fluorescence techniques
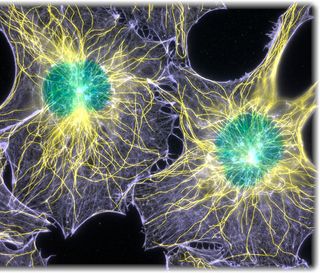
Cell biologists use fluorescence imaging to study cellular structures and processes, often in living cells. Through the staining or tagging of DNA, proteins and other targets with a fluorescent dye, fluorescence imaging techniques have revealed mechanisms of cell division, embryonic development and nerve regeneration. Biologists can now observe whole biological systems with a resolution of about 20 nanometers—2,500 times smaller than the width of a single human hair. Researchers are also working to create new types of dyes, optically active materials, labeling strategies, fluorescent proteins and nanoparticles to produce brighter and more detailed images.
Fluorescence techniques: One new approach to fluorescence imaging uses light to manipulate cell activity. In this video, a blue laser beam activates a protein called Rac1, which then stimulates the movement of a human cancer cell. The technique can turn Rac1—and potentially many other proteins—on and off at exact times and places in living cells. Credit: Yi Wu, the Hahn lab, University of North Carolina.
Live cell imaging
Live cell imaging allows researchers to watch individual cells or subcellular components while barely disrupting the cell. With the aid of high-resolution microscopes, cameras and fluorescent sensors, they can create time-lapse movies of cellular and molecular interactions. What they see can answer questions about gene expression, cell division (mitosis), structural changes during programmed cell death and more.
Sign up for the Live Science daily newsletter now
Get the world’s most fascinating discoveries delivered straight to your inbox.
Live cell imaging: This video of cells from an African clawed frog used live cell imaging to capture abnormal mitosis in action. Just one late-to-align chromosome delays anaphase, the stage when chromosomes segregate to the two ends of the cell. Credit: John Daum and Gary Gorbsky, Oklahoma Medical Research Foundation.
Single-molecule imaging
Biologists use an array of tools to study single molecules, including molecular manipulation with optical tweezers, single-molecule fluorescence spectroscopy, and microscopy and scanning methods to map molecular surfaces. By observing a single molecule, researchers can avoid errors that result from averaging data across many molecules and explore details within complex molecular mixtures.

Electron microscopy
Electron microscopy uses a beam of electrons to illuminate and magnify cells that have been stained and prepared on slides. Electron microscopes can achieve about 1,000 times greater magnification than light-based imaging and are useful for studying any basic cellular process as well as developing molecular tags for genetic studies or live cellimaging. They've been instrumental in finding the exact locations of complex molecular assemblies within cells and in determining the structures of proteins.
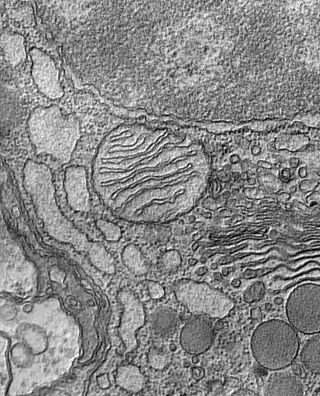
Making sense of imaging data
Scientists use quantitative imaging techniques (which go by names like FRET, FRAP and speckle microscopy) to measure direct molecular interactions inside cells. The data teach them more about how proteins act together to produce cellular functions—something they can’t learn from analyzing individual molecules—as well as how proteins transition between activities. The computers that sort through imaging data are programmed to recognize certain patterns and eliminate background noise, allowing efficient analysis of stacks of images.
Learn more:
This Inside Life Science article was provided to LiveScience in cooperation with the National Institute of General Medical Sciences, part of the National Institutes of Health.

Fake Botox injections have sickened 22, hospitalized 11, CDC warns

Weapons chest found on wreck of 15th-century 'floating castle' sheds light on 'military revolution at sea'

Explosive black hole flare from the center of our galaxy reconstructed from 'a single flickering pixel' using AI and Einstein's equations
